My journey as the 2022 American Academy of Underwater Sciences (AAUS) Mitchell Scientific Diving Intern for the Our World Underwater Scholarship Society (OWUSS) began May 16th as I ventured to East Boothbay, Maine. This summer, I am working at Bigelow Laboratory for Ocean Studies in Dr. Doug Rasher’s Lab where I am assisting Dr. Rasher as well as PhD students Rene Francolini, Dara Yiu and Shane Farrell (2018 Dr. Lee H. Somers AAUS Intern) with a project entitled “Maine-eDNA”. This 5-year project, funded by the National Science Foundation, involves multiple Maine institutions, and aims to improve our understanding of Maine’s coastal ecosystems using molecular ecological tools. As someone who was born and raised in Maine, I was wicked excited to find out I would be participating in such a crucial project, especially one involving the ever-advancing world of eDNA.


For those who do not know, eDNA stands for environmental DNA and this is a relatively new and upcoming molecular tool in ocean sciences and stands to transform how we monitor and understand global ocean ecosystems. An easy way I learned to understand eDNA, is for instance: if you have any furry or hairy pet, you always end up covered in their hair all the time. Anything that “sheds” off your body will have DNA – your entire genetic code. If you take a sample of dust from your floor, you will certainly find lots of DNA belonging to your furry animal, and plenty of human DNA as well. Of course, you don’t need DNA to know about you and your pets, but you may also find a small amount of DNA belonging to insects or rodents – which would show evidence of critters you may not see often but are secretly living in your house. In any body of water, the same process happens. For example, marine animals such as fish shed scales, mucus, or cells into their environment. So, analyzing eDNA – which is, in this instance, all of the DNA collected from a water sample – can be a powerful “forensics” tool to assess who lives in that habitat. Using eDNA is important because it allows scientists to collect information about the total biodiversity in the ecosystem by metabarcoding all the species (fish, algae, invertebrates, microbes etc.) at a location, as well as tell us about organisms that are too rare, small, or hidden to see with our own eyes.
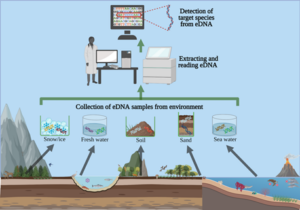
Image by Liam Whitmore, University of Limerick, CC BY-ND (https://theconversation.com/environmental-dna-how-a-tool-used-to-detect-endangered-wildlife-ended-up-helping-fight-the-covid-19-pandemic-158286). Visual explanation demonstrating the flow of eDNA metabarcoding, which starts with the species from an environmental sample to DNA extraction, and results in a “barcode” for the species found in the sample.
The Rasher lab’s project in the larger Maine-eDNA program, is focused on studying “Species on the Move” within kelp forest (rocky reef) ecosystems across the Gulf of Maine (GoM). Our goals are to track changes in species distribution (i.e. the loss of native species and the arrival of new species to the ecosystem), to study the ecological impacts of changing reef communities, and to develop models that help predict these species geographic range shifts. Now you may be wondering, why are the species moving? As a Mainer, I have grown up seeing the impacts of warming in the GoM, but what many people do not know is that the GoM is warming faster than 96.2% of the world’s oceans (GMRI 2021). Additionally, the Gulf of Maine Research Institute (GMRI) recorded the longest marine heat wave ever last year(2021), which lasted from April through most of August. Long story short, species are on the move in the GoM because of ocean warming and marine heat waves, which directly reduces the survival of kelp (a group of cold-water species that create forests) as well as cause the formation of red algae “turf reefs”. Kelp and red algae are quite different – the loss of big, complex structures created by kelp may potentially lead to other changes in the flora and fauna on the rocky reefs across the coast. The transformation of kelp forests to reefs dominated by red algae may have consequences for important commercial species, as their larval and juvenile stages depend on kelp forests as refuge from predators.
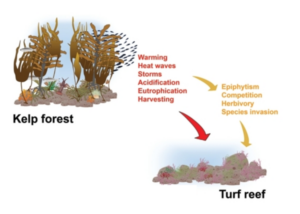
Modified image from Filbee-Dexter and Wernberg in their article, “Rise of Turfs: A New Battlefront for Globally Declining Kelp Forests”. This depicts the direct (red) and indirect (yellow) drivers of a transition from kelp forest to turf reef (Filbee-Dexter and Wernberg 2018).
How are we collecting data to meet the goals of “Species on the Move” project? Through traditional ecological surveys and experiments in conjunction with eDNA analysis. That is where I come in and I get to be in the field collecting data and participating in lab work. As the title of this blog posts suggests, this summer (and science in general) does not involve an everyday 9 to 5 schedule. Instead, our field days are sometimes from 8 am to 11 pm! Each field day consists of going to one of ten study sites. We try our best to pre-pack the boat with gear, otherwise it is packed the morning of, and we try to leave the dock around 9 am.

Pictured above (left) is the Bigelow vessel stern and in the opening of the trees on land is Bigelow Laboratory!

Pictured above (right) includes the PVC frames for squid pops which I’ll talk about below.
On the way to the dive site, we attach line with buoys to PVC frames, because upon arrival to the site all six frames are deployed overboard in a straight line, spaced 10 m apart. Each frame consists of four “squid pops” which are circular cut outs of dried squid. There are two on the top frame to entice fish to get an estimate of predation intensity and two on the bottom frame for invertebrate (e.g., crab, lobster) predation intensity, which we will later compare between sites that have healthy kelp forests to those where kelp has disappeared. Once all the frames are out, we anchor the boat at the GPS location for the dive site and get ready for the dives. Below is a written dive plan that does a great job at explaining what is required at every dive site. I will do my best to explain each dive 🙂
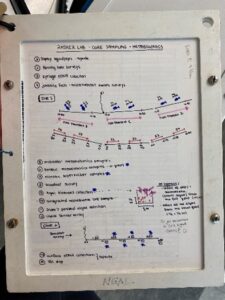

The first dive includes roving fish surveys, eDNA collection (using the syringes pictured above), and juvenile fish and microhabitat swath surveys. Basically, we take two 50 m transects and swim 100 m total, while collecting roving fish data. I also collect four syringes (totaling 2L) at two locations along the first transect and repeat the same process on the second transect. Then on the way back to the starting point, I assist Dara with juvenile fish swaths by spotting tiny fish for 15 m increments along the transect. All the above is repeated on the third and last for the last 50 m transect.

Me and my eDNA syringes in a kelp forest in northern Maine.
The second dive includes conducting eight quadrat surveys along a 50 m transect. Each quadrat survey includes assessment of percent cover of kelp and other algae found within the 1 m2 PVC frame, stipe counts of brown algae, counts of fish, as well as counts and percent cover of invertebrates (e.g., sponges, barnacles, etc.). In addition, within some of these replicate quadrats we collect metabolomic water samples and collections of microbial communities as part of Shane’s effort to understand how the loss of kelp forests impacts the chemical and microbial microenvironments of the reef. After Shane and Dara take estimates of algae cover, count animals, and collect water, I am responsible for harvesting and collecting all the algae within six quadrats, so that we can calculate an estimate of biomass. This involves collecting all kelp found in the full 1 m2 quadrat as well as collecting all other algae in a 0.25 m2 area of quadrat by hand. By collecting the kelps and algae’s it allows us to get precise measurements of the relative abundances of kelp and red algae species – and ID all the cryptic red algae species – which is important for tracking “species on the move” and for eDNA comparison. Some algae species must be viewed under a microscope in the lab or sent off to a facility to be genetically barcoded, to reveal their identity.
The last dive is used to finish the last quadrat survey, but most likely to collect any leftover gear or more algae.

Left to Right: Me, Dara, and Shane before we entered the water for our third dive of the day!
After the dives, we collect the squid pop frames and head back to Bigelow, but the fun for the day does not end there. Once we get back to the lab, take everything off the boat, and clean/rinse gear, lab work starts! First, all the eDNA water samples are put through a filter and all the DNA from the water sample is then stuck to a piece of filter paper, which we save for analysis later.

Seawater from eDNA syringe in graduated cylinder is poured into the filter seen in background.

Filter paper with DNA from filtered seawater collected from Allen Island.
The last activity of a dive day is sorting, IDing, and weighing all the different algae collected from the quadrats! I took a phycology class my sophomore year of college, but I missed out on the lab portion due to COVID. So, this has been a great experience to apply what knowledge I have and of course learn more about algae! I have become familiar with many of the brown algae like Agarum and Laminaria, green algae like Chaetomorpha and Ulva sp, and red blade algae like Chondrus, Porphyra, Lomentaria, Palmaria, and Euthora. These species I have become very familiar with and I am able to identify them underwater too!
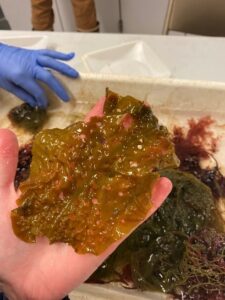
Agarum! Known for its holes which is believed to be an adaptation for fast moving water environments.

Lomentaria! Looks like a cactus 🙂
The filamentous branched and branched red tubes are more difficult to ID by just looking at them, so we usually examine them under the microscope. Dara has been a great resource for algae ID and she typically asks me what I think the algae is based on characteristics rather than telling me what the algae is under the scope. Some characteristics that are important for filamentous algae ID include cortication around the cells and pericentral cells.

Algae sorting!
So far, we have completed our spring survey at 10 dive sites, that range from turf reefs in the south to lush kelp forests in the north. For the following few weeks, I will assist the lab with some molecular work, learn about the process of preparing DNA samples for sequencing, and then prepare for the late summer round of diving. I am eager to share with everyone what I learn in the lab!

